
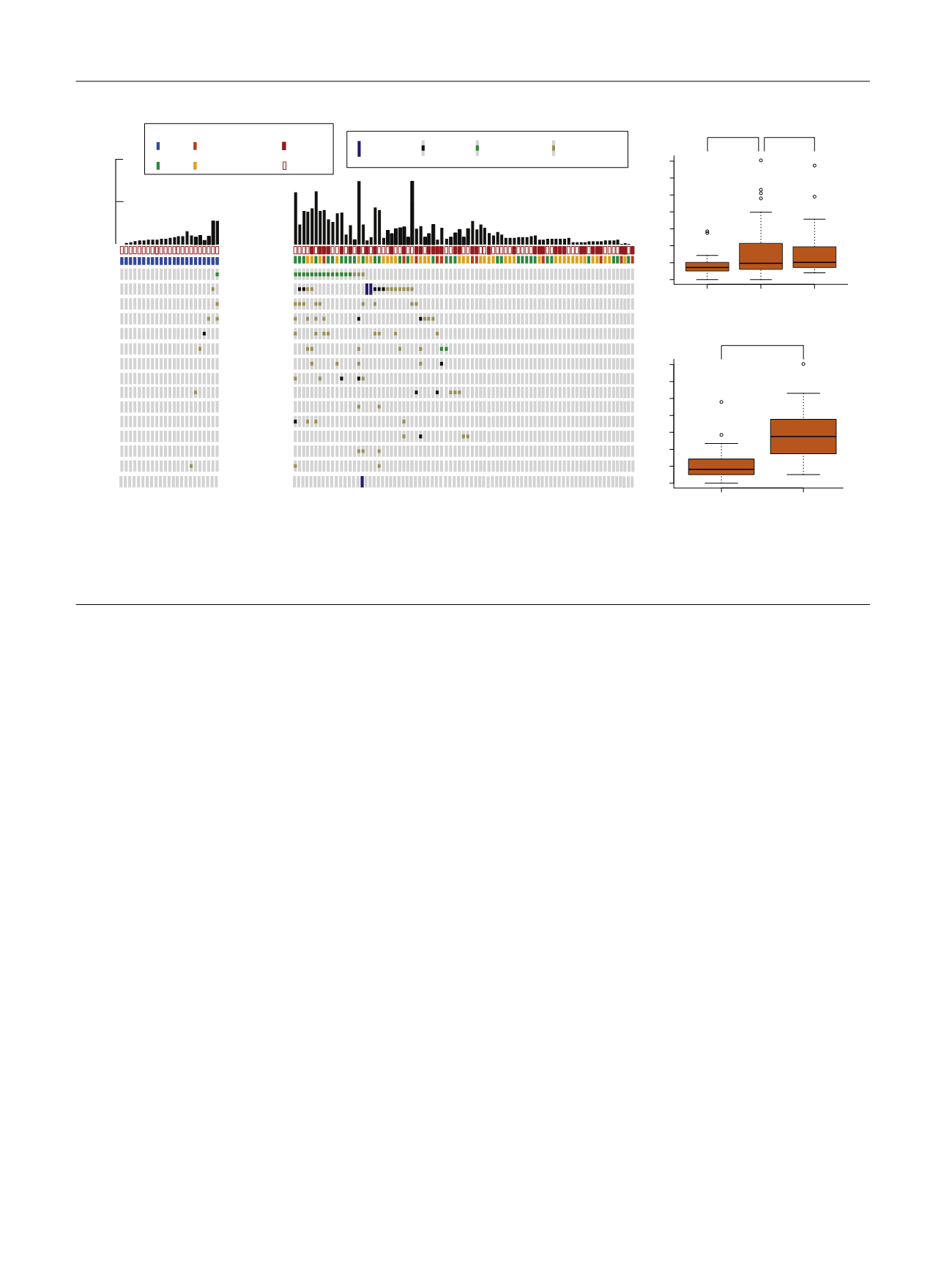
found to cluster around conserved helicase domains
(Supplementary Fig. 9). Less frequent DDR gene alterations
were also observed in
ATM, BRCA1, BRCA2, ERCC4, PALB2,
CHECK2, FANCC
, and
MSH6
( Fig. 2 A). One LGTa with five total
mutations had a truncating
POLE
H1901Lfs*15 mutation
that is likely not functionally relevant given its location near
the 3’ end of the protein and the absence of a hypermutation
phenotype. The remaining DDR gene alterations in NMIBC
were missense variants of unknown significance.
Since MIBC has one of the largest mutational burdens of
all tumor types studied by TCGA, we used mutational
burden per megabase on targeted exon sequencing to infer
associations between mutational burden and clinical
variables
[21] .High-grade NMIBC tumors had a larger
median mutational burden per megabase than low-grade
NMIBC (9, interquartile rage [IQR]: 6–21 vs 7, IQR: 5–10,
p
= 0.032) and had a mutational burden similar to MIBC (10,
IQR: 7–19,
p
= 0.84;
Fig. 2B). Furthermore, tumors with
deleterious DDR gene alterations had a significantly higher
mutational burden as compared with tumors with intact
DDR genes (26, IQR: 15–36 vs 8, IQR: 5–12,
p
<
0.001;
Fig. 2 C, Supplementary Fig. 10). Similarly,
ERCC2
-mutated
tumors had a significantly higher mutational burden than
ERCC2
wild-type tumors (29, IQR: 23–37 vs 8, IQR: 6–14,
p
<
0.001; Supplementary Fig. 11).
3.5.
Genomic correlates with recurrence after BCG therapy
With the objective of identifying genomic alterations
associated with recurrence after BCG, we examined
62 patients within the NMIBC cohort who had high-grade
disease and were treated with a 6-wk induction course of
BCG without maintenance therapy. Tumor recurrence
occurred in 52% (32/62) of patients at a median follow-
up of 24 mo. When evaluating all genes in the 341-gene
panel that were altered in at least five tumors in the
62-patient cohort, only
ARID1A
mutations were signifi-
cantly associated with an increased risk of recurrence after
BCG treatment (hazard ratio [HR] = 3.14, 95% confidence
interval [CI]: 1.51–6.51,
p
= 0.002;
Table 2 , Fig. 3 ). This held
true even when correcting for multiple comparisons
(
p
= 0.04) and when
ARID1A
missensemutations of unknown
significance were included (HR = 3.08, 95% CI: 1.49–6.35,
p
= 0.002;
Table 2 ,Supplementary Fig. 12).
ARID1A
muta-
tions were also associated with tumor recurrence within the
larger cohort of 100 patients managed by TUR with or
without adjuvant intravesical therapy (HR = 2.07, 95% CI:
1.10–3.88,
p
= 0.024). No significant associations were
identified between the examined clinical variables and
recurrence free survival in either the 62 patient BCG cohort
or the larger 100 patient cohort (Supplementary Table 8).
As reports associating
TP53
and treatment outcomes in
NMIBC are conflicting
[22] ,we examined recurrence free
survival in
TP53
-altered tumors alone and in combination
with
MDM2
and found no association with recurrence after
BCG (
p
= 0.94 and
p
= 0.36, respectively; Supplementary Fig.
13). Furthermore, tumors with alterations in
ERBB2
and
FGFR3
had similar recurrence rates after BCG as wild type
tumors (
p
= 0.3), further supporting adjuvant trials of
targeted inhibitors of these kinases (Supplementary Fig. 7B).
[(Fig._2)TD$FIG]
Low-grade NMIBC (
n
= 23)
High-grade NMIBC (
n
= 82)
ERCC2
ATM
ATR
BRCA2
POLE
BRCA1
ERCC4
PALB2
CHEK2
ERCC5
FANCC
MSH6
MSH2
FANCA
MRE11A
Grade/Stage
HG Tis
HG Ta HG T1
LG Ta
Mutation count on MSK-IMPACT
100
50
Grade/Stage
Carcinoma in situ
Carcinoma in situ
Yes
No
Deep deletion
Recurrent
Missense mutation
Truncating
Mutation
Novel
Missense mutation
DDR Unaltered
DDR Altered
0 10 20 30 40 50 60 70
Mutational burden per MB
p
< 0.001
Low-grade
NMIBC
MIBC
0 10 20 30 40 50 60 70
Mutational burden per MB
High-grade
NMIBC
p
= 0.032
p
= 0.84
A
B
C
Fig. 2 – (A) A comparison of DNA damage repair gene alterations found in high and low grade NMIBC. (B) Box and whisker plot comparing mutational
burden per megabase between low-grade NMIBC (
n
= 23), high-grade NMIBC (
n
= 82), and MIBC (
n
= 40). (C) Box and whisker plot comparing
mutational burden per megabase between high-grade NMIBC tumors with altered and unaltered DNA damage repair genes (
n
= 82).
DDR = DNA damage repair; MB = megabase; MIBC = muscle invasive bladder cancer; MSK-IMPACT = Memorial Sloan Kettering Cancer Center-Integrated
Mutation Profiling of Actionable Cancer Targets; NMIBC = nonmuscle invasive bladder cancer.
E U R O P E A N U R O L O G Y 7 2 ( 2 0 1 7 ) 9 5 2 – 9 5 9
956
















